Blogs & News
Text Mining Clinical Histopathology Reports
The Aridhia team has an ongoing commitment to researching and integrating emerging technologies and innovations into our product, so we encourage staff to collaborate on Research and Development projects that can support the translation of ideas into reality. This allowed us to deliver an initial investigation into the use of text mining and natural language processing (NLP) techniques for clinical information systems. This particular study focused primarily on breast and lung cancer histopathology reports using the in-house domain experience in this field to drive development.
The potential reach of text mining in clinical practice is an area of growing interest, as there remains a significant proportion of unstructured data that’s currently underutilised for making inferences. We wanted to get our people thinking more about the subject and how we could help our clients to get more out of the vast amounts of unstructured data in their clinical systems.
Text Mining Frameworks
From a technical perspective, Aridhia’s Platform & Product Development team has been looking at the current state of the art in terms of existing frameworks for text mining. The investigation began by producing our own rules-based prototype application for performing Named Entity Recognition, which also used the Part-Of-Speech (POS) tagger from the Open NLP. POS tagging is the process of identifying the role of a word in a sentence, i.e. is the word a noun, adjective or noun phrase? The application showed that a rules-based approach can perform well for the semi-structured data, but a larger document corpus will be required for evaluating accuracy. The possibility of using the prototype for performing sentence level and document level classification through supervised machine learning was also investigated.
The study has paused recently at GATE (General Architecture for Text Engineering) and Apache UIMA (Unstructured Information Management Architecture), two well established Java-based frameworks in the field of text analysis. GATE is an active project at the University of Sheffield and during the last 15 years has become widely adopted in various fields. Similarly, UIMA is a well-established project, available as Apache Open source software having initially begun as an IBM project.
Both GATE and UIMA adopt a modular approach to developing text mining applications. The ability to separate the overall tasks into smaller components is central to the philosophy. GATE, for example, is divided into modules termed Processing Resources (PRs) that can operate on a document of interest such as a pathology report. GATE comes with ready-made modules for performing common tasks associated with text mining. These come under the ANNIE project for information extraction. ANNIE also relies on JAPE, an interesting rules language for annotating documents. Figure 1 below explains some typical components relevant to extracting information from text.
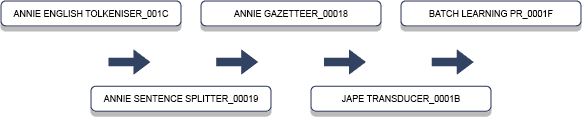
This pipeline uses a word tokeniser to split a document into its component words, so to form a list. This is followed by a sentence boundary detector, not necessarily a simple task as word abbreviations and numbers are just two quick examples where a full-stop alone is not reliable in recognising sentence endings. Therefore, depending on the specific use case, these components need to be customised to suit the requirements based on the style of the text.
Next in the pipeline is an ANNIE Gazetteer and JAPE Transducer, both of which allow domain keywords or groups of words to be recognised and annotated in the document. Finally, the Batch Learning module is GATE’s component for applying machine learning.
Machine learning is an area that uses statistical techniques to adapt, based on previously seen documents. A typical project will use what’s known as a training data set to achieve this, and apply “knowledge” gained from this training data to a separate test data set, in order to evaluate performance and prepare the system for production.
The utility of text mining solutions to a problem will depend on the nature of the task and the requirements in terms of accuracy. GATE allows both rule-based and machine learning approaches to be taken, as well as tools to assess the performance of the application.
Text mining for clinical unstructured data is an exciting area, particularly in relation to the DECIPHER Health project. In terms of clinical practice some key areas of relevance will include the following:
- Detecting the recurrence of a tumour.
- Correlation of SNOMED coding and bottom line diagnosis.
- Allowing the identification of new prognostic indicators (e.g. immunohistochemistry markers).
- Identifying the number of patients with specific genetic profiles suitable for drug trials.
- Percentage of resections that have been adequately excised (resection margins).
- How many of the resections are re-excisions.
Additionally, for radiologists the accuracy of radiologically guided biopsies is of interest, as is the general correlation between pathology and radiology reports.
If you are interested in more detail on the project, please get in touch.
APPENDIX: REFERENCES
See the user guide for more information.
Further reading: Getting More Out of Biomedical Documents with GATE’s Full Lifecycle Open Source Text Analytics, by Hamish Cunningham, Valentin Tablan, Angus Roberts, Kalina Bontcheva (Computation Biology).
Aridhia is not responsible for the content of external links.
July 16, 2013
daniel
Dan joined Aridhia in 2012 and brings over 10 years’ experience in software development and data management. He is a keen reader around the biological sciences and is close to completing a Postgraduate Certificate in Digital Biology from the University of Manchester. Dan has played an active role in Aridhia’s advancement into genomics, developing software and working with various types of NGS data. He also holds a BSc. (Hons) in Biochemistry with Applied Molecular Biology from UMIST, and an MSc. in Intellectual Property Management.
