Blogs & News
Unlocking Global Proteomics Collaboration: How Aridhia Enabled the GNPC
Looking to the future of biomedical research and the opportunities at hand with growing digitalization and democratization of data, a theme begins to emerge:
New and meaningful insights – impacting real lives for the better – will come from coupling global data sources and expertise with scalable, secure, and collaborative digital infrastructure.
That vision came to life through our partnership with the AD Data Initiative and the Global Neurodegeneration Proteomics Consortium (GNPC) – a groundbreaking international initiative that set out to build the world’s largest harmonized proteomics dataset for neurodegenerative diseases.1
Led by Gates Ventures and Janssen R&D (Johnson & Johnson), and supported by a consortium of 23 global cohorts, the GNPC represents a new model of open, collaborative science. And we’re proud that the Aridhia Digital Research Environment (DRE) provided the HITRUST Workspaces as the secure, cloud-native foundation to make it possible.
The Challenge: Scale, Sensitivity, and Sovereignty
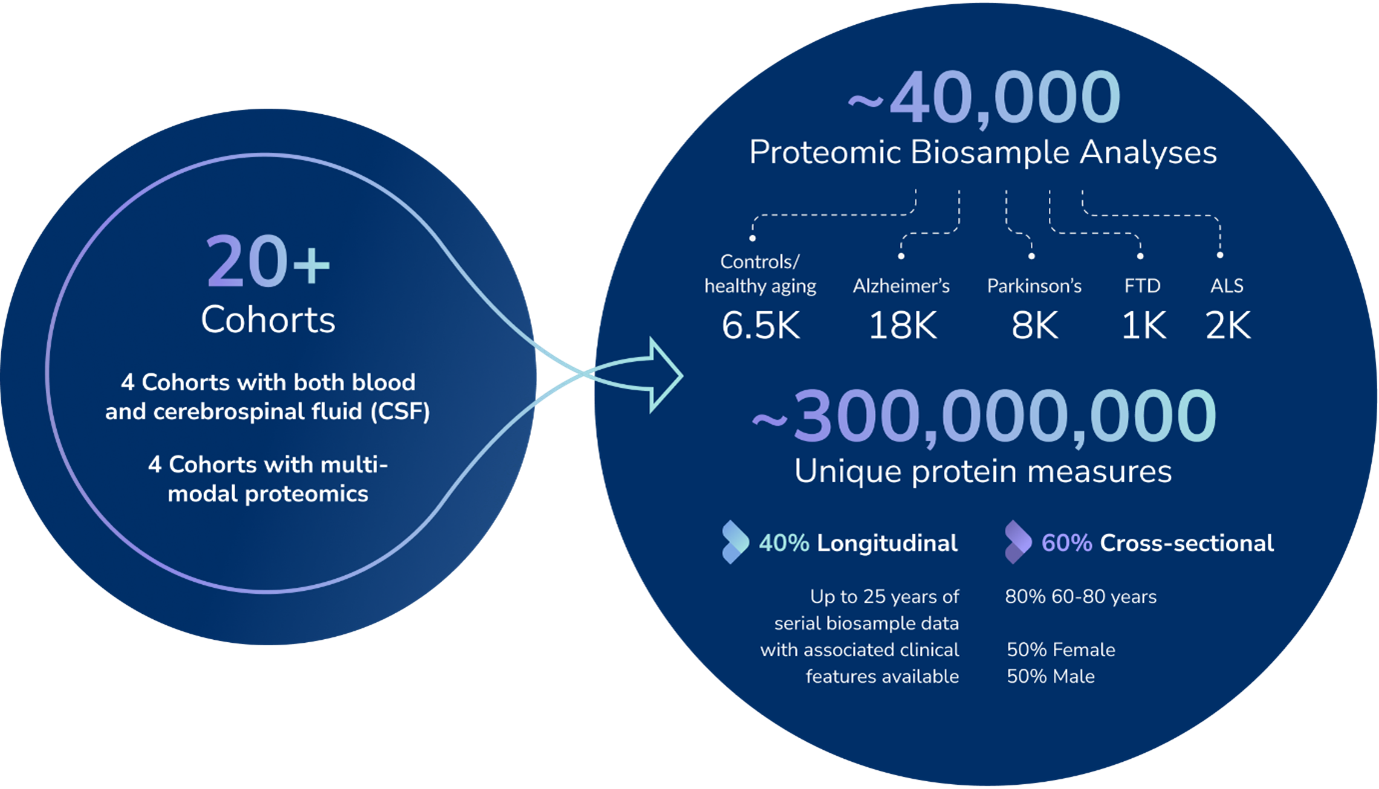
Neurodegenerative diseases such as Alzheimer’s, Parkinson’s, ALS, and frontotemporal dementia (FTD) are some of the most complex and devastating health challenges of our time. Understanding their underlying mechanisms requires studying thousands of protein biomarkers across vast, diverse populations.
To accelerate this, GNPC brought together more than 35,000 biosamples from over 18,000 individuals, across dozens of global research institutions. These samples were analyzed on cutting-edge platforms like SomaScan, Olink, and mass spectrometry, generating more than 250 million unique protein measurements.
But such scale introduced major challenges:
This is where the Aridhia DRE came in…
The Solution: A Secure Network of Workspaces in the DRE
Rather than attempting to centralize data in a single location, GNPC adopted a federated model using the Aridhia DRE as a network of secure, cloud-based Workspaces. Each Workspace was tailored to a specific stage of the data lifecycle – collection, curation, harmonization, or analysis – enabling modular, auditable collaboration without raw data ever leaving approved Workspaces or jurisdictions.
Contributor Workspaces
Each participating cohort was provisioned a private Workspace within the DRE. These acted as secure digital silos, where raw proteomic and clinical data could be uploaded, cleaned, and documented by the original data custodians. No sensitive data was ever moved outside these workspaces, ensuring full control and compliance.
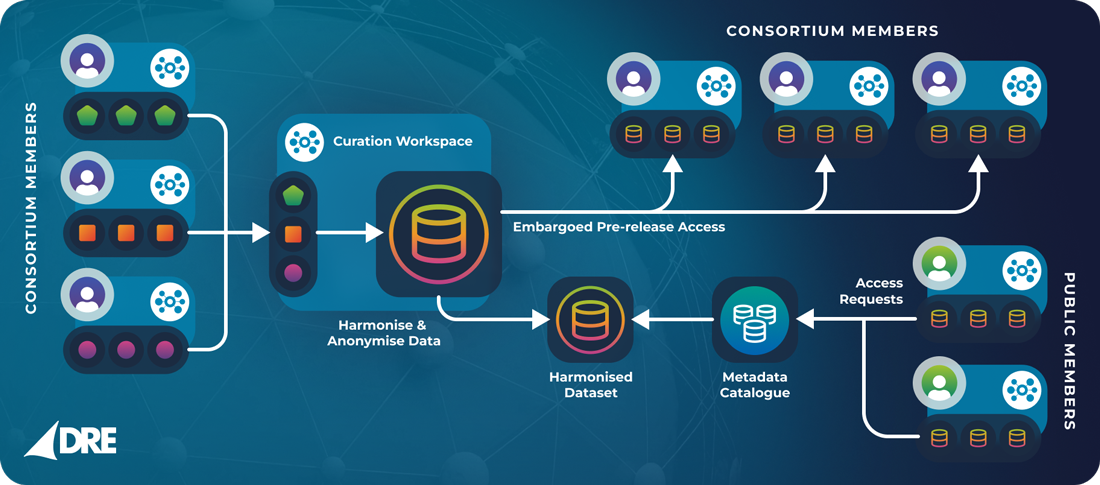
Curation and Harmonization Hubs
Dedicated workspaces were assigned to GNPC’s curation team. Here, data engineers used embedded tools like R Development Environment, Python, SQL, and Jupyter Notebooks to perform:
From there, harmonization teams integrated curated datasets across cohorts and platforms. The DRE’s native support for version control, lineage tracking, and reproducibility allowed teams to trace each protein or clinical variable back to its source – a critical component of scientific transparency and downstream validation.
Analysis Workspaces
Once harmonized, data was made available to approved researchers through analysis-ready workspaces. These environments were fully equipped for statistical and machine learning workflows, with curated libraries and documentation to guide cross-disease analyses.
Researchers could explore questions like:
All of this was done in situ, without data being downloaded or moved. Results could be exported only after audit approval, preserving GNPC’s high bar for security and reproducibility.
Collaboration at Global Scale
In many ways, the GNPC represents the kind of digitally native science the DRE was built for:
What made it all work was the DRE’s shared digital infrastructure that respected local data governance while enabling cross-border collaboration.
“The Aridhia DRE allowed us to turn a global network of proteomics data into a living, collaborative research platform. HITRUST Workspaces gave us the ability to curate, harmonize, and analyze tens of millions of proteomic data points across multiple diseases. Without it, harmonizing sensitive datasets across jurisdictions while maintaining security and compliance would have been nearly impossible.”
Dr. Farhad Imam, GNPC Co-Lead
The DRE allowed researchers to operate as a distributed team with real-time access to tools, documentation, and harmonized data. With the platform’s high security standards and Workspaces designed for flexibility, curators and researchers alike were able to effectively and collaboratively do their work without ever compromising security.
Looking Ahead: A Blueprint for Open Biomedical Science
In July 2025, GNPC released Version 1 of its Harmonized Data Set (HDS V1) to the global research community via the AD Workbench, built on top of the Aridhia DRE. It’s already fueling new discoveries, including:
As GNPC evolves toward its second data release, the DRE serves as a backbone enabling rapid iteration, data expansion, and equitable access.
“The GNPC proves that international data science can be both collaborative and secure. By using the DRE powered Workspaces to build a network of controlled-access Workspaces, the consortium was able to move fast, stay compliant, and unlock critical insights into neurodegenerative diseases.”
Dr. Niranjan Bose, AD Data Initiative Interim Executive Director
V1 of the HDS included samples from the US, Germany, the Netherlands, Sweden, Spain, and the UK. V2 aims to bring in at least 5000 samples from diverse cohorts in Central and South America, Asia, and Africa for better global representation.
We’re honored to support a project that not only advances neurodegeneration research, but also sets a new standard for secure, collaborative, data-driven discovery.
August 22, 2025
Kara Lasater
Kara joined Aridhia in 2020 and leads the Customer team as Head of Customer Success where she works closely with clients to deliver valuable solutions on the DRE. Kara leads with a commitment to understanding and addressing customer needs and fostering a customer-centric culture at Aridhia. In her free time, she enjoys hiking and taking road trips with her family and labradoodle, Cosmo.