Blogs & News
Nonlinear Mixed Effect Modelling – The use of nlmixr2 in a DRE workspace
Nonlinear mixed effect modelling is a core component of many model-informed drug development (MIDD) approaches pursued by industry sponsors seeking to develop new drugs, vaccines or devices. Likewise, it is a necessary skill for regulatory authorities who must review such models and judge the credibility of recommendations made by sponsors based on using such models. It is often also the backbone for many decision support systems or model-informed precision dosing (MIPD) approaches implemented to individualize and hopefully optimize patient pharmacotherapy.
Historically, much of the industry and regulatory experience with this approach has been based on the NONMEM algorithm (a software package developed by Stuart L. Beal and Lewis B. Sheiner in the late 1970s at University of California, San Francisco). NONMEM is now in the category of commercial software for which annual license fees are required. While a few commercial software alternatives have been developed, these still require annual and group license agreements and incur financial and personnel costs. An alternative to these solutions has been the development of open science solutions which are generally supported by the original developer in collaboration with the open science community.
The nlmixr algorithm
In the case of nonlinear mixed effect modelling, the most well-known and established open science solution is the nlmixr and now nlmixr2 algorithm. nlmixr is an R package for fitting general dynamic models, pharmacokinetic (PK) models and pharmacokinetic-pharmacodynamic (PKPD) models in particular, with either individual data or population data with a long list of developers and a broad community of users.
As of this writing the website tabulated over 41,000 total downloads with > 270 downloads per month. Given the longstanding and continued use of the solution there is growing confidence in the algorithm’s performance but also confidence in being able to trust its output for regulatory submissions (as with new drug and/or vaccine development) or as part of a precision dosing strategy to optimize pharmacotherapy to individual patients. In both situations, considerations for patient privacy, security, access to analysis-ready data and audit trails can complement the algorithm’s performance with a holistic implementation that increases the overall confidence and fidelity of the solution.
nlmixr in the DRE
For initial exploratory analysis of datasets, Aridhia’s Digital Research Environment (DRE) provides Data Table Analytics modules (pre-built analysis modules designed in R). These serve as a useful jumping off point to concentrate the focus of proposed models.
nlmixr2 being used in RStudio inside a DRE workspace.
Ease of use, cost, and access are obviously factors to consider for any research tool, and these are additional areas in which the DRE concentrates its efforts. The route for getting up and running with nlmixr2 on AWS for example, is a lengthy process where you must first provision and configure a virtual machine instance, installing and further configuring R and RStudio, before then installing the relevant packages and beginning your work.
In a DRE workspace, not only do you have access to RStudio as a containerized application, it requires no virtual machine instance to be created at all, reducing resource costs. The DRE has always been positioned as a multidisciplinary environment; data scientists should not have to act as an IT or Ops department configuring VMs instead of focusing on the work suited to them. Installing nlmixr2 (one line of code (install.packages("nlmixr2")) and a 30 second installation) is the only barrier to beginning your analysis. All outputs and reports can be generated within the workspace, ready for approved export at a later time.
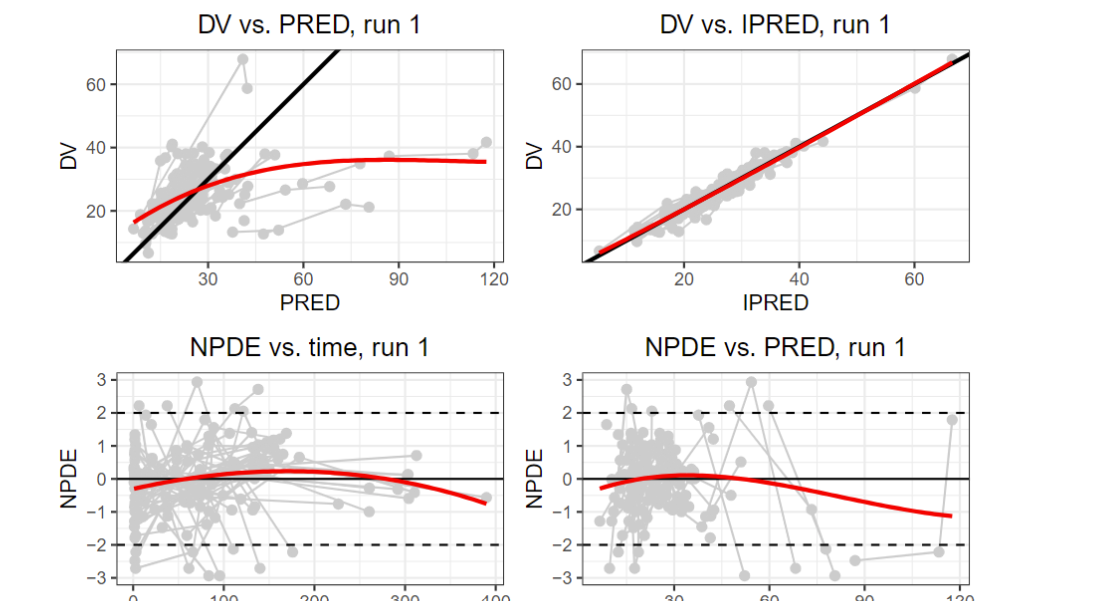
Plots generated in a DRE workspace using the nlmixr2 algorithm.
Add to this the comprehensive audit and versioning capabilities that come out of the box. We have previously covered the in-built Git feature; as patient privacy concerns are a key driver here, the Git repository is held securely inside the workspace. It can only be removed from the workspace file system via the secure airlock.
Given all of these considerations, the marriage of nlmixr and the Aridhia DRE is one where obvious synergies are formed and the implementation of the solution in a DRE workspace can improve the overall useability and performance of a key, open science solution for the application of both MIDD and MIPD approaches.
(n.b. in related reading, you can find Jeff’s paper here on an AI Approach to Generating MIDD Assets Across the Drug Development Continuum)
Authors
 Dr. Jeff Barrett is the Chief Science Officer at Aridhia promoting healthcare and life science partners to collaborate, access and share secure data to deliver better patient outcomes. Before Aridhia, he was Senior Vice-President at the Critical Path Institute serving as the Executive Director of the Rare Disease Cures Accelerator, Data Analytics Platform. Jeff was previously Head of Quantitative Sciences at the Bill & Melinda Gates Medical Research Institute. Prior to MRI, he was Vice President, of Translational Informatics at Sanofi Pharmaceuticals. Jeff spent 10+ years at the University of Pennsylvania where he was Professor, Paediatrics and Director, Laboratory for Applied PK/PD at the Children’s Hospital of Philadelphia.
Dr. Jeff Barrett is the Chief Science Officer at Aridhia promoting healthcare and life science partners to collaborate, access and share secure data to deliver better patient outcomes. Before Aridhia, he was Senior Vice-President at the Critical Path Institute serving as the Executive Director of the Rare Disease Cures Accelerator, Data Analytics Platform. Jeff was previously Head of Quantitative Sciences at the Bill & Melinda Gates Medical Research Institute. Prior to MRI, he was Vice President, of Translational Informatics at Sanofi Pharmaceuticals. Jeff spent 10+ years at the University of Pennsylvania where he was Professor, Paediatrics and Director, Laboratory for Applied PK/PD at the Children’s Hospital of Philadelphia.
–
 Janani Murthy joined Aridhia in August 2021 as a Software Developer/Data Engineer. She has a background in computer science engineering with over 10 years of experience in software development in her previous organisations, having completed her MSc in Big Data Tools and Technologies at Glasgow Caledonian University. She is part of the Customer Engineering team and provides technical solutions, implementation and support for the data science lifecycle within the DRE.
Janani Murthy joined Aridhia in August 2021 as a Software Developer/Data Engineer. She has a background in computer science engineering with over 10 years of experience in software development in her previous organisations, having completed her MSc in Big Data Tools and Technologies at Glasgow Caledonian University. She is part of the Customer Engineering team and provides technical solutions, implementation and support for the data science lifecycle within the DRE.
April 11, 2024
Jeff Barrett
Dr. Jeff Barrett is the Chief Science Officer at Aridhia promoting healthcare and life science partners to collaborate, access and share secure data to deliver better patient outcomes. Before Aridhia, he was Senior Vice-President at the Critical Path Institute serving as the Executive Director of the Rare Disease Cures Accelerator, Data Analytics Platform. Jeff was previously Head of Quantitative Sciences at the Bill & Melinda Gates Medical Research Institute. Prior to MRI, he was Vice President, of Translational Informatics at Sanofi Pharmaceuticals. Jeff spent 10+ years at the University of Pennsylvania where he was Professor, Paediatrics and Director, Laboratory for Applied PK/PD at the Children’s Hospital of Philadelphia.
